Note
Go to the end to download the full example code
Spatial properties of brain regions
When a reference space is specified, brain regions can expose a range of spatial properties.
We start by selecting an atlas and a space
import siibra
atlas = siibra.atlases.get("human")
# print the spaces defined in the atlas
print(atlas.spaces)
InstanceTable:
- BIGBRAIN_MICROSCOPIC_TEMPLATE_HISTOLOGY
- FREESURFER_FSAVERAGE
- MNI_152_ICBM_2009C_NONLINEAR_ASYMMETRIC
- MNI_COLIN_27
Choose a space
space = atlas.spaces.get("icbm 2009c asym")
print(space.name)
# and a region
v1_left = atlas.get_region("v1 left", parcellation='julich 2.9')
print(v1_left)
MNI 152 ICBM 2009c Nonlinear Asymmetric
Area hOc1 (V1, 17, CalcS) left
The Region.spatial_props() method computes the centroid and volume of a brain region. Note that a brain region might in general consist of multiple separated components in the space. Also note that in siibra, spatial properties are always represented in millimeter units of the physical coordinate system of the reference space, not in voxel units.
SpatialProp(cog=None, components=[SpatialPropCmpt(centroid=<Point((-8.515510862347028, -82.38838819846315, 2.00540745659805), space='minds/core/referencespace/v1.0.0/dafcffc5-4826-4bf1-8ff6-46b8a31ff8e2', sigma_mm=0.0)>, volume=10541.0)], space=<Space(identifier='minds/core/referencespace/v1.0.0/dafcffc5-4826-4bf1-8ff6-46b8a31ff8e2', name='MNI 152 ICBM 2009c Nonlinear Asymmetric', species='Homo sapiens')>)
The returned centroid is siibra.Point object. Such spatial primitives are covered in more detail in Locations in reference spaces. For now, we just acknowledge that this minimizes misinterpretation of the coordinates, since a siibra Point is explicitly linked to its space.
centroid = props.components[0].centroid
print(centroid)
centroid.space.name
Point in MNI 152 ICBM 2009c Nonlinear Asymmetric [-8.515510862347028,-82.38838819846315,2.00540745659805]
'MNI 152 ICBM 2009c Nonlinear Asymmetric'
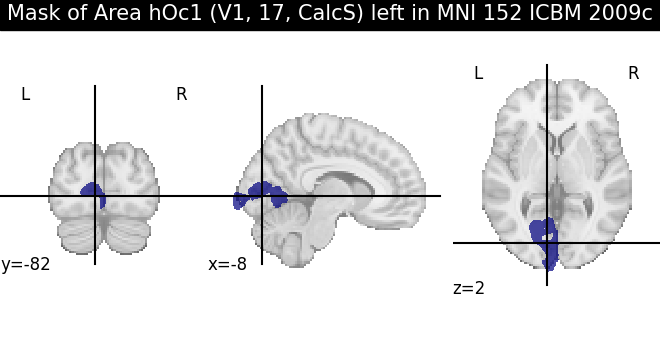
We can also generate a binary mask of the region in a given space and maptype, which gives us a Nifti1Image object as provided by nibabel, and which we can directly visualize using plotting functions like the ones in nilearn:
mask = v1_left.get_regional_map(space, maptype="labelled")
from nilearn import plotting
plotting.plot_roi(mask.fetch(), title=f"Mask of {v1_left.name} in {space.name}")
<nilearn.plotting.displays._slicers.OrthoSlicer object at 0x7f72a9184250>
Total running time of the script: (2 minutes 8.057 seconds)