Note
Go to the end to download the full example code
Assigning coordinates to brain regions
siibra can use statistical parcellations maps to make a probabilistic assignment of exact and imprecise coordinates to brain regions. We start by selecting the Julich-Brain probabilistic maps from the human atlas, which we will use for the assignment.
import siibra
from nilearn import plotting
Choose a parcellation map. We demonstrate the use of probabilistic maps here.
with siibra.QUIET: # suppress progress output
julich_pmaps = siibra.get_map(
parcellation="julich 2.9",
space="mni152",
maptype="statistical"
)
Assigning exact coordinate specifications.
We now determine the probability values of cytoarchitectonic regions at a an
arbitrary point in MNI reference space. The example point is manually chosen.
It should be located in PostCG of the right hemisphere. For more information
about the siibra.Point class see Locations in reference spaces. The output is a pandas
dataframe, which includes the values of different probability maps at the
given location. We can sort the table by these values, to see that the region
with highest probability is indeed the expected region.
point = siibra.Point((27.75, -32.0, 63.725), space='mni152')
with siibra.QUIET: # suppress progress output
assignments = julich_pmaps.assign(point)
assignments.sort_values(by=['map value'], ascending=False)
Assigning coordinate specifications with location uncertainty.
Typically, coordinate specifications are not exact. For example, we obtain a
position from an sEEG electrode, which has several millimeters of uncertainty
(especially after warping it to standard space!). If we specify the position
with a location uncertainty, siibra will not just read out the values of
the probability maps, but instead generate a 3D Gaussian blob with a
corresponding standard deviation, and correlate the 3D blob with the maps. We
then obtain the weighted average of the map values over the blob, but also
additional measures of comparison: A correlation coefficient, the intersection
over union (IoU), a containedness score of the blob wrt. the region
(“contained”), and a containness score of the region wrt. the blob
(“contains”). Per default, the resulting table is sorted by correlation
coefficient. Here, we query the assignments with a containedness score of at
least 0.5, that is, the regions in which the uncertain point is likely
contained.
point_uncertain = siibra.Point((27.75, -32.0, 63.725), space='mni152', sigma_mm=3.)
with siibra.QUIET: # suppress progress output
assignments = julich_pmaps.assign(point_uncertain)
assignments.query('`input containedness` >= 0.5').dropna(axis=1)
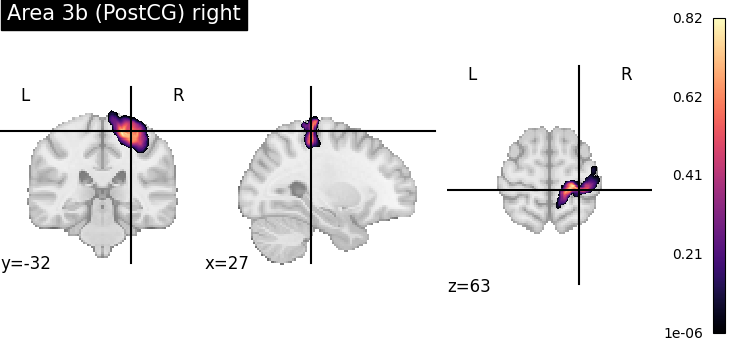
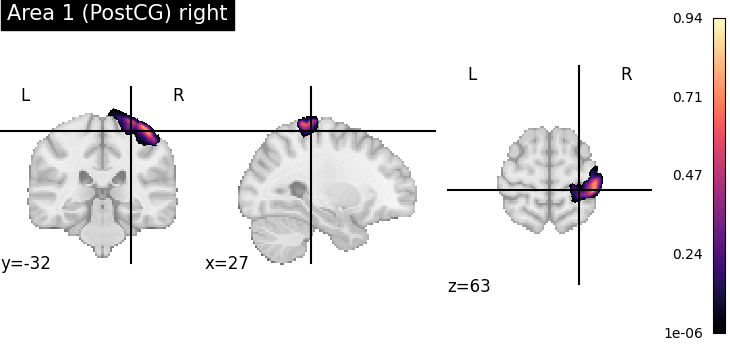
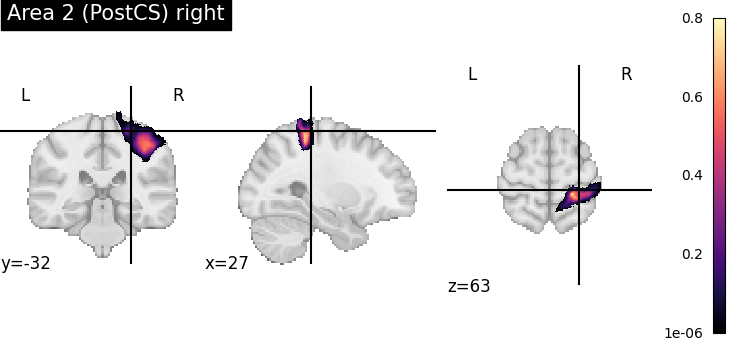
To verify the result, we plot the assigned probability maps at the requested position.
for index, assignment in assignments[assignments['input containedness'] >= 0.5].iterrows():
pmap = julich_pmaps.fetch(region=assignment['region'])
plotting.plot_stat_map(pmap, cut_coords=tuple(point), title=assignment['region'], cmap='magma')
Total running time of the script: (0 minutes 22.571 seconds)