Note
Go to the end to download the full example code
Adding a custom parcellation map
Sometimes you might want to use a custom parcellation map to perform feature queries in siibra. For this example, we retrieve a freely available AICHA - Atlas of Intrinsic Connectivity of Homotopic Areas (Tzourio-Mazoyer N, Landeau B, Papathanassiou D, Crivello F, Etard O, Delcroix N, Mazoyer B, Joliot M (2002) Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. Neuroimage 15:273-289.). This atlas provided in the MNI ICBM 152 space.
import siibra
from nilearn import plotting
Load the custom parcellation map from the online resource. For the retrieval, siibra’s zipfile connector is very helpful. We can use the resulting NIfTI to create a custom parcellation map inside siibra.
# connect to the online zip file
conn = siibra.retrieval.ZipfileConnector(
"http://www.gin.cnrs.fr/wp-content/uploads/aicha_v1.zip"
)
# the NIfTI file is easily retrieved:
nifti = conn.get("AICHA/AICHA.nii")
# and create a volume on space MNI152 (note that this assumes our
# external knowledge that the map is in MNI152 space)
volume = siibra.volumes.from_nifti(nifti, 'mni152', "AICHA")
# The text file with label mappings has a custom format. We provide a tsv
# decoder to extract the list of region/label pairs since the txt file is tab
# seperated.
volume_info = conn.get("AICHA/AICHA_vol1.txt", decode_func=siibra.retrieval.requests.DECODERS['.tsv'])
volume_info
Now we use this to add a custom map to siibra.
regionnames = [
name.replace('-R', ' right').replace('-L', ' left')
for name in volume_info['nom_l']
]
labels = [int(label) for label in volume_info['color']]
custom_map = siibra.volumes.parcellationmap.from_volume(
name="AICHA - Atlas of Intrinsic Connectivity of Homotopic Areas",
volume=volume,
regionnames=regionnames,
regionlabels=labels
)
let’s plot the final map
plotting.plot_roi(custom_map.fetch())
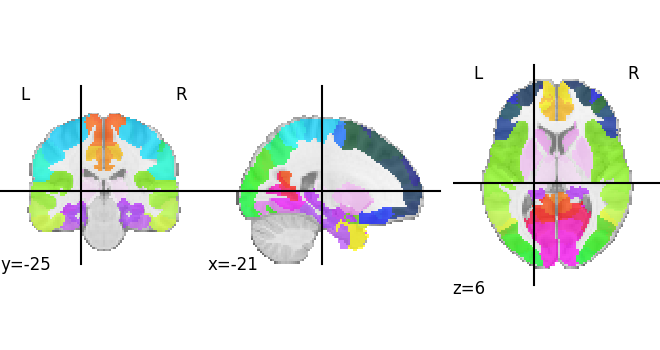
<nilearn.plotting.displays._slicers.OrthoSlicer object at 0x7f72af0d4100>
We can already use this map to find spatial features, such as BigBrain intensity profiles, gene expressions, volume of interests, sections…
region = custom_map.parcellation.get_region('S_Rolando-1 left')
profiles = siibra.features.get(
region,
siibra.features.cellular.BigBrainIntensityProfile
)[0]
print(f"{len(profiles)} intensity profiles found.")
2146 intensity profiles found.
On the other hand, some features are only anchored to a sementic region object and the link to the custome region is not directly known to siibra. However, siibra circumvents this by comparing volumes of these regions to assign a link between them.
volume = region.get_regional_map('mni152')
receptor_density = siibra.features.get(
volume,
siibra.features.molecular.ReceptorDensityFingerprint
)
print(f"{len(receptor_density)} layerwise cell density found.")
# The relation of volume to the anatomical anchor is provided in
# `anchor.last_match_description`
for d in receptor_density:
print(d.anchor.last_match_description)
6 layerwise cell density found.
'Volume Subtree mask built from Area 44 (IFG) in space MNI 152 ICBM 2009c Nonlinear Asymmetric' overlaps with 'Volume Mask of S_Rolando-1 left in AICHA Atlas of Intrinsic Connectivity of Homotopic Areas on MNI 152 ICBM 2009c Nonlinear Asymmetric in space MNI 152 ICBM 2009c Nonlinear Asymmetric'
'Volume Subtree mask built from Area PFt (IPL) in space MNI 152 ICBM 2009c Nonlinear Asymmetric' overlaps with 'Volume Mask of S_Rolando-1 left in AICHA Atlas of Intrinsic Connectivity of Homotopic Areas on MNI 152 ICBM 2009c Nonlinear Asymmetric in space MNI 152 ICBM 2009c Nonlinear Asymmetric'
'Volume Subtree mask built from Area 3b (PostCG) in space MNI 152 ICBM 2009c Nonlinear Asymmetric' overlaps with 'Volume Mask of S_Rolando-1 left in AICHA Atlas of Intrinsic Connectivity of Homotopic Areas on MNI 152 ICBM 2009c Nonlinear Asymmetric in space MNI 152 ICBM 2009c Nonlinear Asymmetric'
'Volume Subtree mask built from Area 4p (PreCG) in space MNI 152 ICBM 2009c Nonlinear Asymmetric' overlaps with 'Volume Mask of S_Rolando-1 left in AICHA Atlas of Intrinsic Connectivity of Homotopic Areas on MNI 152 ICBM 2009c Nonlinear Asymmetric in space MNI 152 ICBM 2009c Nonlinear Asymmetric'
'Volume Subtree mask built from Area 44 (IFG) in space MNI 152 ICBM 2009c Nonlinear Asymmetric' overlaps with 'Volume Mask of S_Rolando-1 left in AICHA Atlas of Intrinsic Connectivity of Homotopic Areas on MNI 152 ICBM 2009c Nonlinear Asymmetric in space MNI 152 ICBM 2009c Nonlinear Asymmetric'
'Volume Subtree mask built from Area PFop (IPL) in space MNI 152 ICBM 2009c Nonlinear Asymmetric' overlaps with 'Volume Mask of S_Rolando-1 left in AICHA Atlas of Intrinsic Connectivity of Homotopic Areas on MNI 152 ICBM 2009c Nonlinear Asymmetric in space MNI 152 ICBM 2009c Nonlinear Asymmetric'
To get a more detailed assignment scores, one can make use of map assignment discussed in Anatomical Assignment, see Assign modes in activation maps to brain regions.
Total running time of the script: (5 minutes 36.958 seconds)