Note
Go to the end to download the full example code
Neurotransmitter receptor densities
EBRAINS provides transmitter receptor density measurements linked to a selection of cytoarchitectonic brain regions
in the human brain (Palomero-Gallagher, Amunts, Zilles et al.). These can be accessed by calling the
siibra.features.get() method with feature types in siibra.features.molecular modality, and by
specifying a cytoarchitectonic region. Receptor densities come as cortical profiles and regional fingerprints,
both tabular style data features.
import siibra
If we query this modality for the whole atlas instead of a particular brain region, all linked receptor density features will be returned.
parcellation = siibra.parcellations.get('julich 2.9')
all_features = siibra.features.get(parcellation, siibra.features.molecular.ReceptorDensityFingerprint)
print("Receptor density fingerprints found at the following anatomical anchors:")
print("\n".join(str(f.anchor) for f in all_features))
Receptor density fingerprints found at the following anatomical anchors:
CA1 (Hippocampus)
CA2 (Hippocampus)
CA3 (Hippocampus)
Area 44 (IFG)
CA2 (Hippocampus)
Area TE 2.1 (STG)
CA1 (Hippocampus)
CA2 (Hippocampus)
CA3 (Hippocampus)
CA1 (Hippocampus)
CA2 (Hippocampus)
CA3 (Hippocampus)
Area PGp (IPL)
CA3 (Hippocampus)
Area PFt (IPL)
CA2 (Hippocampus)
Area 45 (IFG)
Area FG1 (FusG)
CA1 (Hippocampus)
CA2 (Hippocampus)
CA3 (Hippocampus)
Area PFcm (IPL)
Area TE 1.0 (HESCHL)
CA3 (Hippocampus)
Area hOc2 (V2, 18)
CA1 (Hippocampus)
Area 7A (SPL)
Area 3b (PostCG)
Area hOc2 (V2, 18)
Area PF (IPL)
Area FG2 (FusG)
Area PGa (IPL)
Area 4p (PreCG)
CA1 (Hippocampus)
Area hOc3d (Cuneus)
Area 44 (IFG)
Area hOc3v (LingG)
Area PFm (IPL)
Area PFop (IPL)
DG (Hippocampus)
Area hOc1 (V1, 17, CalcS)
DG (Hippocampus)
When providing a particular region instead, the returned list is filtered accordingly. So we can directly retrieve densities for the primary visual cortex:
v1_fingerprints = siibra.features.get(
siibra.get_region('julich 2.9', 'v1'),
siibra.features.molecular.ReceptorDensityFingerprint
)
v1_fingerprints[0].plot()
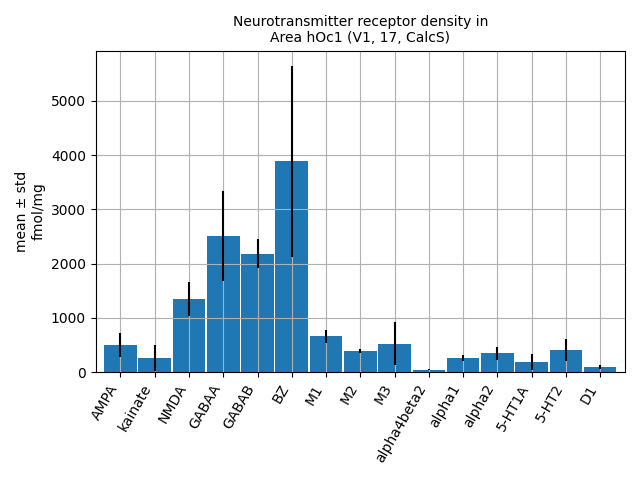
<Axes: title={'center': 'Neurotransmitter receptor density in\nArea hOc1 (V1, 17, CalcS)'}, ylabel='mean ± std\nfmol/mg'>
Each feature includes a data structure for the fingerprint, with mean and standard values for different receptors. The following table thus gives us the same values as shown in the polar plot above:
v1_fingerprints[0].data
Many of the receptor features also provide a profile of density measurements at different cortical depths, resolving the change of distribution from the white matter towards the pial surface. The profile is stored as a dictionary of density measures from 0 to 100% cortical depth.
v1_profiles = siibra.features.get(
siibra.get_region('julich 2.9', 'v1'),
siibra.features.molecular.ReceptorDensityProfile
)[0]
for p in v1_profiles:
print(p.receptor)
if "GABAA" in p.receptor:
print(p.receptor)
break
print(p.data)
p.plot()
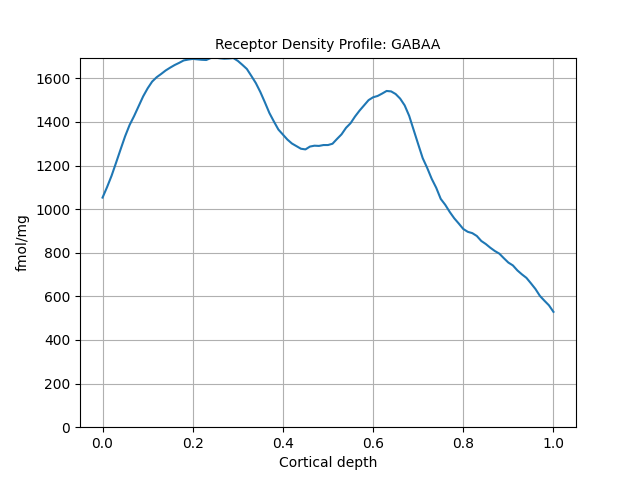
5-HT1A
5-HT2
alpha1
alpha2
alpha4beta2
AMPA
BZ
D1
GABAA
GABAA
Receptor density (fmol/mg)
0.00 1053
0.01 1101
0.02 1153
0.03 1213
0.04 1274
... ...
0.96 634
0.97 602
0.98 580
0.99 559
1.00 529
[101 rows x 1 columns]
<Axes: title={'center': 'Receptor Density Profile: GABAA'}, xlabel='Cortical depth', ylabel='fmol/mg'>
Total running time of the script: (0 minutes 13.900 seconds)