Note
Go to the end to download the full example code
Comparative analysis of brain organisation in two brain regions
siibra data features simplify analysis of multimodal aspects of brain regions. In this example, we select a region from the Broca region in the inferior frontal gyrus, IFG 44, involved in language processing, and a region from the visual system in the occipital cortex, V1.
import siibra
import matplotlib.pyplot as plt
We start by selecting the two region objects from the parcellation, using simple keyword specifications.
Next, we choose a set of feature modalities that we’re interested in. We start with “fingerprint” style features for cell and receptor densities, which provide average measurements across multiple samples, and can be easily compared visually.
modalities = [
siibra.features.molecular.ReceptorDensityFingerprint,
siibra.features.cellular.LayerwiseCellDensity,
siibra.features.cellular.LayerwiseBigBrainIntensities,
]
We iterate the regions and modalities to generate a grid plot.
fig, axs = plt.subplots(len(modalities), len(regions))
ymax = [4500, 150, 30000]
for i, region in enumerate(regions):
for j, modality in enumerate(modalities):
features = siibra.features.get(region, modality)
print(region, modality, len(features))
if len(features) > 0:
fp = features[-1]
fp.plot(ax=axs[j, i])
axs[j, i].set_ylim(0, ymax[j])
fig.tight_layout()
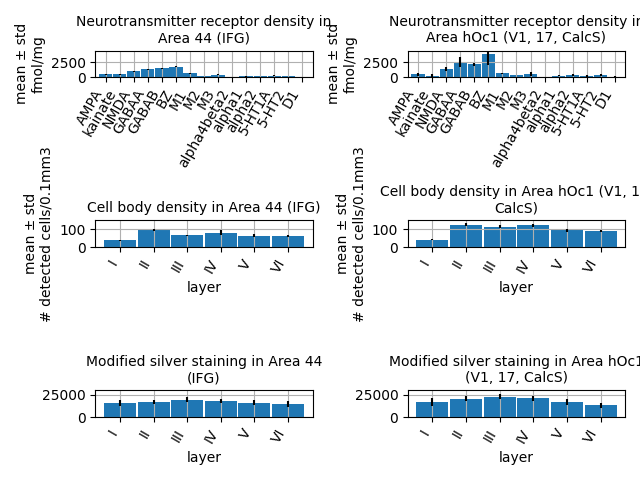
Area 44 (IFG) <class 'siibra.features.tabular.receptor_density_fingerprint.ReceptorDensityFingerprint'> 2
Area 44 (IFG) <class 'siibra.features.tabular.layerwise_cell_density.LayerwiseCellDensity'> 1
Area 44 (IFG) <class 'siibra.features.tabular.layerwise_bigbrain_intensities.LayerwiseBigBrainIntensities'> 1
Area hOc1 (V1, 17, CalcS) <class 'siibra.features.tabular.receptor_density_fingerprint.ReceptorDensityFingerprint'> 1
Area hOc1 (V1, 17, CalcS) <class 'siibra.features.tabular.layerwise_cell_density.LayerwiseCellDensity'> 1
Area hOc1 (V1, 17, CalcS) <class 'siibra.features.tabular.layerwise_bigbrain_intensities.LayerwiseBigBrainIntensities'> 1
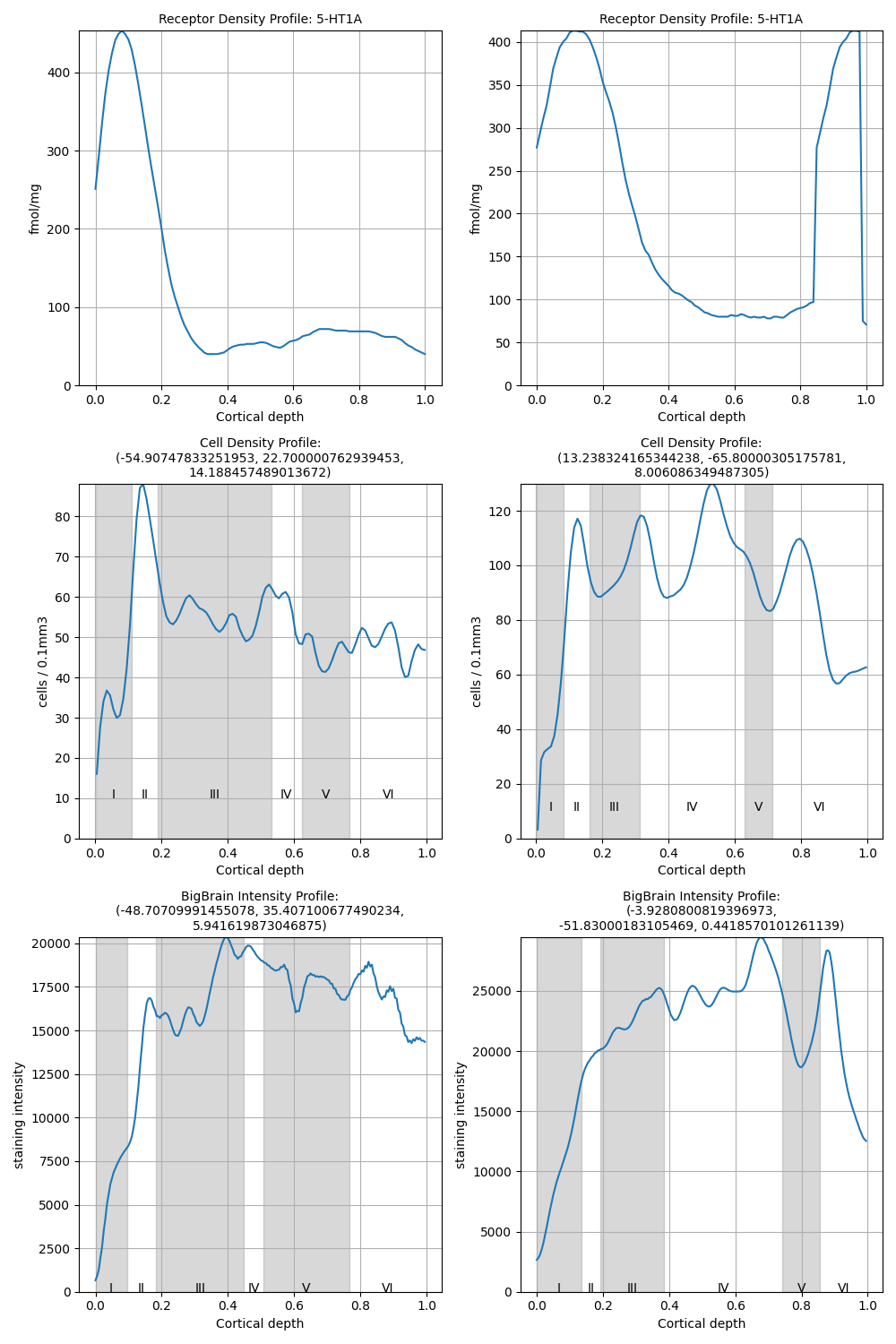
For the same measurement types, we can also sample individual cortical profiles, showing density distributions from the pial surface to the gray/white matter boundary in individual tissue samples. Cortical profiles queries form Compound features for easy handling of the profiles. We can browse through the elements with integer index. Let us select the first ones for comparison.
modalities = [
siibra.features.molecular.ReceptorDensityProfile,
siibra.features.cellular.CellDensityProfile,
siibra.features.cellular.BigBrainIntensityProfile,
]
fig, axs = plt.subplots(len(modalities), len(regions))
for i, region in enumerate(regions):
for j, modality in enumerate(modalities):
cf = siibra.features.get(region, modality)[0]
p = cf[0]
p.plot(ax=axs[j, i])
fig.set(figheight=15, figwidth=10)
fig.tight_layout()
Total running time of the script: (2 minutes 21.172 seconds)