Note
Go to the end to download the full example code
Parcellation-based functional data
siibra provides access to parcellation-averaged functional data such as blood-oxygen-level-dependent (BOLD) signals.
import siibra
We start by selecting an atlas parcellation.
julich_brain = siibra.parcellations.get("julich 2.9")
The tables are queried as expected, using siibra.features.get, passing the parcellation as a concept. Here, we query for regional BOLD signals. Since a single query may yield hundreds of signal tables for different subjects of a cohort and paradigms, siibra groups them as elements into Compound features. Let us select “rfMRI_REST1_LR_BOLD” paradigm.
features = siibra.features.get(julich_brain, siibra.features.functional.RegionalBOLD)
for f in features:
print(f.name)
if f.paradigm == "rfMRI_REST1_LR_BOLD":
cf = f
print(f"Selected: {cf.name}'\n'" + cf.description)
200 Regional BOLD features cohort: HCP, paradigm: rfMRI_REST1_LR_BOLD
Selected: 200 Regional BOLD features cohort: HCP, paradigm: rfMRI_REST1_LR_BOLD'
'Resting-state functional connectivity (FC) is frequently used to predict behavioral, clinical and demographic subject traits. This type of brain connectome can be derived from blood-oxygen-level-dependent (BOLD) signals that reflect the activation of individual brain regions parcellated according to a given brain atlas. Deriving FC from BOLD signals typically involves the estimation of the amount of synchronized coactivations between the BOLD time series of different brain regions. However, several measures of synchronization exist and which one of these metrics is suited best may deviate from study to study. In parallel, the appropriate selection of the brain parcellation is nowadays also still an open issue. This dataset hence comprises the region-based BOLD signals extracted from the resting-state functional magnetic resonance imaging (fMRI) data of 200 healthy subjects included in the Human Connectome Project. The time series were extracted for 20 different state-of-the-art parcellations. The neuroimaging community may use the data of this repository to study, for example, how different measures of synchronization affect the resting-state FC under various parcellation conditions.
200 Regional BOLD features cohort: HCP, paradigm: rfMRI_REST1_RL_BOLD
200 Regional BOLD features cohort: HCP, paradigm: rfMRI_REST2_LR_BOLD
200 Regional BOLD features cohort: HCP, paradigm: rfMRI_REST2_RL_BOLD
We can select a specific element by integer index
print(cf[0].name)
print(cf[0].subject) # Subjects are encoded via anonymized ids
000 - Regional BOLD cohort: HCP, paradigm: rfMRI_REST1_LR_BOLD
000
The signal data is provided as pandas DataFrames with region objects as columns and indices as as a timeseries.
time
0 days 00:00:00 7218.786845
0 days 00:00:00.720000 7186.080235
0 days 00:00:01.440000 7196.478754
0 days 00:00:02.160000 7162.358546
0 days 00:00:02.880000 7173.673591
...
0 days 00:14:20.400000 7206.168284
0 days 00:14:21.120000 7210.908622
0 days 00:14:21.840000 7187.603819
0 days 00:14:22.560000 7204.503983
0 days 00:14:23.280000 7203.507897
Name: Area hOc3v (LingG) left, Length: 1200, dtype: float64
We can visualize the signal strength per region by time via a carpet plot. In fact, plot_carpet method can take a list of regions to display the data for selected regions only.
selected_regions = [
'SF (Amygdala) left', 'SF (Amygdala) right', 'Area Ph2 (PhG) left',
'Area Ph2 (PhG) right', 'Area Fo4 (OFC) left', 'Area Fo4 (OFC) right',
'Area 7A (SPL) left', 'Area 7A (SPL) right', 'CA1 (Hippocampus) left',
'CA1 (Hippocampus) right', 'CA1 (Hippocampus) left', 'CA1 (Hippocampus) right'
]
cf[0].plot_carpet(regions=selected_regions)
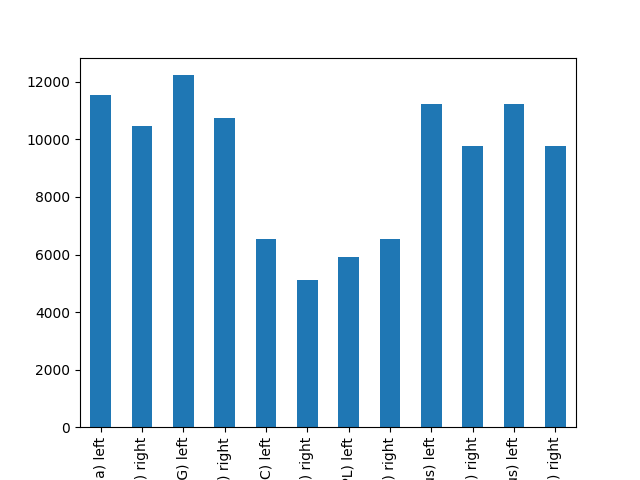
Alternatively, we can visualize the mean signal strength per region:
cf[0].plot(regions=selected_regions)
<Axes: >
Total running time of the script: (0 minutes 54.440 seconds)