Note
Go to the end to download the full example code
Gene expressions
siibra can query gene expression data from the Allen brain atlas. The gene
expressions are linked to atlas regions by coordinates of their probes in MNI
space. When querying feature by a region, siibra automatically builds a
region mask to filter the probes.
Hint
This feature is used by the JuGEx toolbox, which provides an implementation for differential gene expression analysis between two different brain regions as proposed by Bludau et al.
import siibra
from nilearn import plotting
We select a brain region and query for expression levels of GABARAPL2.
<siibra.features.tabular.gene_expression.GeneExpressions object at 0x7f72ad7056d0>
Since gene expressions are spatial features, let’s check the reference space of the results.
space = features[0].anchor.space
print(space)
MNI 152 ICBM 2009c Nonlinear Asymmetric
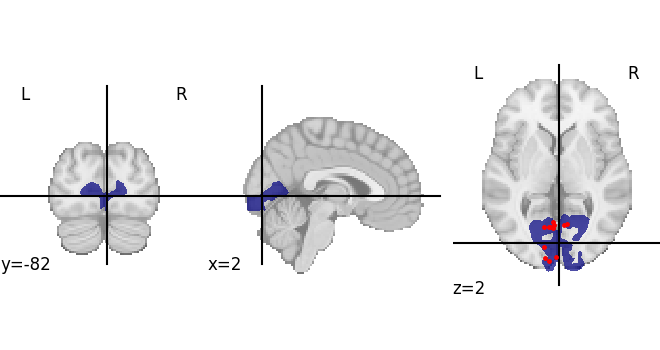
Plot the locations of the probes that were found, together with the region mask of V1.
all_coords = [p.coordinate for p in features[0].anchor.location]
mask = region.get_regional_map(space)
display = plotting.plot_roi(mask.fetch())
display.add_markers(all_coords, marker_size=5)
Total running time of the script: (0 minutes 9.818 seconds)